dna reverse complement multiple sequences
Reverse Complement converts a DNA sequence into its reverse complement or reverse-complement counterpart. If there can be multiple-character insertions go with the other answers.

Is There Multiple Alignment Software That Can See The Reverse Complement
G with C.

. Then reversing the resulting sequence. The entire IUPAC DNA alphabet is supported and the case of each input sequence character is maintained. The get method of a dictionary allows you to specify a default value if the key is not in the dictionary.
17 rows Reverse andor complement DNA sequences. Or getting the complement and then reversing. Enter your DNA sequence in the.
Sequence to reverse complement. As a double helix made of two complementary strands a DNA fragment can be sequenced as two. Alternatively sequence IDs can be entered in the constraint box to select specific sequences.
Click the Find Repeats element and set the options of the repeats search algorithm. Show a Translated CDS in 3-Letter Format. This program generates the complement or reverse complement of a DNA sequence.
Creating complement of DNA sequence and reversing it C In other words it is reverse complement of a DNA sequence which can be easily achieved by reversing the DNA sequence and then getting its complement. You may want to work with the reverse-complement of a sequence if it contains an ORF on the reverse strand. T with A.
Reverse Complement Reverse Complement converts a DNA sequence into its reverse complement or reverse-complement counterpart. Reverse complement DNA sequences. Paste the raw or FASTA sequence into the text area below.
You may want to work with the reverse-complement of a sequence if it contains an ORF on the reverse strand. Reverse complement of DNA strand using Python. Click File Save to save the sequence file in the reverse orientation.
The sequence or sequences to be reverse complemented can be selected in the top window. Creating a MSA aware of the Reverse and Forward Directions. The entire IUPAC DNA alphabet is supported and the case of each input sequence character is maintained.
Most MSA tools are not aware of the input sequences directionality. An example is shown below. Ambiguity codes of the three possible nucleotides are converted as.
3 Sort the levels of genetic material organization from lower to higher level Differentiate DNA from RNA Describe the causes and consequences of the directionality in nucleic acids Identify the reverse complement and reverse complement based on the original sequence Explain why thermophilic organisms have a higher GC content Define semi-conservative DNA replication. Replaced by their complement M K R Y S S V B W W H D N N and the gap - and hard masking letters are unchanged. Convert the DNA sequence into its reverse.
Upload a sequence file in FASTA format. As a preconditioning step I would map all your non ATGC bases to single letters or punctuation or numbers or anything that wont show up in your sequence then reverse the sequence then. The reverse complement of a sequence is formed by exchanging all of its nucleobases with their base complements and then reversing the resulting sequence.
To display the reverse complement of a dsDNA sequence and associated features click View Flip Sequence. A seq TCGGGCCC reverse_complement joincomplementgetbase base for base in reversedseq. IUPAC ambiguity codes of the two possible nucleotides are converted as following.
Open the file datacmdlinefind-repeatsuwl from the UGENE installation directory. Reverse Complement Reverse Complement converts a DNA sequence into its reverse complement or reverse-complement counterpart. Click the Write Sequence element and choose the output file name.
You may want to work with the reverse-complement of a sequence if it contains an ORF on the reverse strand. Therefore if the SAME sequence is used as input into an MSA in different directions a tool like MAFFT automatically outputs an MSA showing that the two sequences are far from each other as we saw above. As DNA sequencing technologies keep improving in scale and cost there is a growing need to develop machine learning models to analyze DNA sequences eg to decipher regulatory signals from DNA fragments bound by a particular protein of interest.
The reverse complement of a DNA sequence is formed by exchanging all instances of. Reverse andor complement DNA. Click the Read Sequence element and choose files with the input sequences.
In general a generator expression is simpler than the original code and avoids creating extra list objects. Value An object of the same class and length as the original object. Input must be single or mutiple sequences in FASTA format.
ReverseComplementxis equivalent to reversecomplementxbut is faster and more memory efficient. C with G. Both DNA and RNA sequence is converted into reverse-complementing sequence of DNA.
This program reverse complements DNA sequences. A with T. TARGET SEQUENCE Paste your DNA sequence below OPTIONS Transformation Options Reverse and Complement Reverse.
Thursday March 3rd 2022. RY KM S and W unchanged. To reverse complement a single stranded sequence see Convert Between Single-Stranded and Double-Stranded Formats.
DNA Sequence Reverse and Complement Online Tool With this tool you can reverse a DNA sequence complement a DNA sequence or reverse and complement a DNA sequence Supports IUPAC ambiguous DNA characters. Hold down the Ctrl button when selecting multiple sequences.

Reverse Complement Polymerase Chain Reaction Wikipedia

Cause Of Failed Dna Sequencing Using Reverse Primer

Reverse Complement Polymerase Chain Reaction Wikipedia
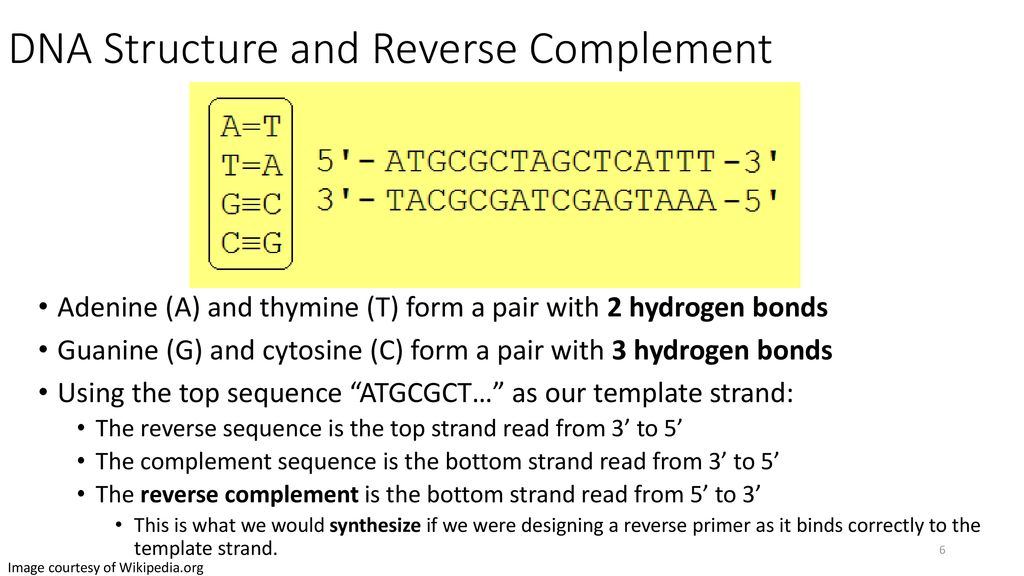
4 29 19 Monday Night Meeting Ben Maldonato Ppt Download

Forward And Reverse Dna Sequence Editing And Alignment Youtube

Sequence Alignment Software Codoncode Aligner

Pairwise Alignment Tutorial Geneious Prime
How To Create A Reverse Complement Sequence

How To Find The Reverse Complement Of A Dna Sequence 2 Steps

How To Find The Reverse Complement Of A Dna Sequence 2 Steps

Reverse Complement Pcr A Novel One Step Pcr System For Typing Highly Degraded Dna For Human Identification Forensic Science International Genetics
Sequence Editing Dna Sequencing Software Sequencher From Gene Codes Corporation

How To Find The Reverse Complement Of A Dna Sequence 2 Steps

Multiple Dna Sequence Alignment Of Consensus Binding Sites Of Penr Download Scientific Diagram

2 An Example Of 5 Coding And 3 Reverse Complementary Strand Primers Download Scientific Diagram
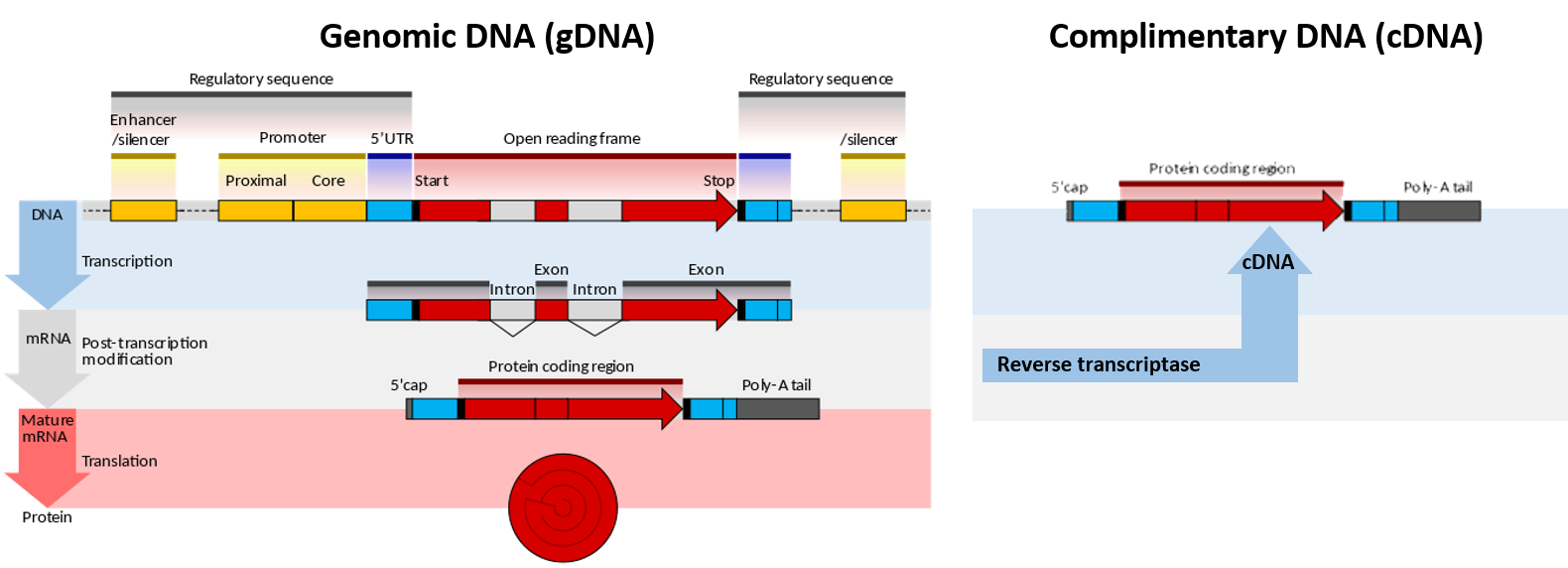
Chapter 5 Investigating Dna Chemistry

How To Find The Reverse Complement Of A Dna Sequence 2 Steps
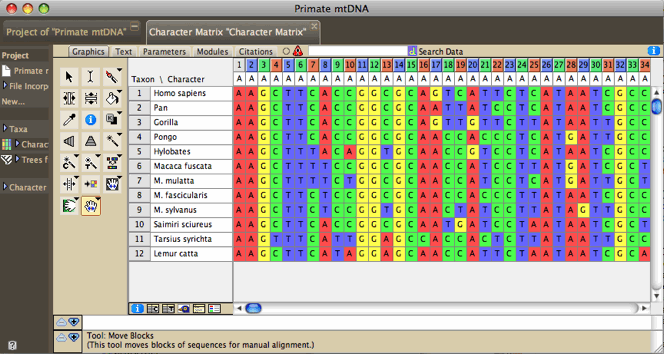
Comments
Post a Comment